Sparse multi-task lasso
Table of Contents
Introduction
UTMOST (Hu et al. 2019) fits predictive models for gene expression across multiple conditions jointly, based on optimizing the penalized objective:
\[ \newcommand\mb{\mathbf{B}} \newcommand\me{\mathbf{E}} \newcommand\mr{\mathbf{R}} \newcommand\mx{\mathbf{X}} \newcommand\my{\mathbf{Y}} \ell = \frac{1}{2} \sum_{k=1}^m \Vert \my_{\cdot k} - \mx \mb_{\cdot k} \Vert_F + \lambda_1 \sum_{k=1}^m \Vert \mb_{\cdot k} \Vert + \lambda_2 \sum_{j=1}^p \Vert \mb_{j \cdot} \Vert_2 \]
This approach was previously proposed by Lee et al. 2010. We reimplemented
this method in the package gtexpred. Here, we evaluate our implementation
on simulations.
Setup
import functools as ft import hyperopt as hp import mtlasso import numpy as np import pandas as pd import sklearn.linear_model as sklm import sklearn.metrics as skm import sklearn.model_selection as skms import timeit import txpred
import rpy2.robjects.packages import rpy2.robjects.pandas2ri rpy2.robjects.pandas2ri.activate()
%matplotlib inline %config InlineBackend.figure_formats = set(['retina'])
import colorcet import matplotlib import matplotlib.pyplot as plt plt.rcParams['figure.facecolor'] = 'w' plt.rcParams['font.family'] = 'Nimbus Sans'
Methods
Idealized simulation
def _simulate(X, B, pve): n = X.shape[0] m = B.shape[1] G = X.dot(B) E = np.random.normal(size=(n, m), scale=np.sqrt((1 / pve - 1) * G.var(axis=0))) Y = G + E # Center the data X -= X.mean(axis=0) Y -= Y.mean(axis=0) return X, Y, B def simulate_identical_effects(n, p, m, n_causal, pve, seed): np.random.seed(seed) X = np.random.normal(size=(n, p)) b = np.zeros((p, 1)) b[:n_causal] = np.random.normal(size=(n_causal, 1)) B = np.outer(b, np.ones(m)) return _simulate(X, B, pve) def simulate_shared_effects(n, p, m, n_causal, pve, seed): np.random.seed(seed) X = np.random.normal(size=(n, p)) B = np.zeros((p, m)) B[:n_causal,:] = np.random.normal(size=(n_causal, m)) return _simulate(X, B, pve)
Results
Running time
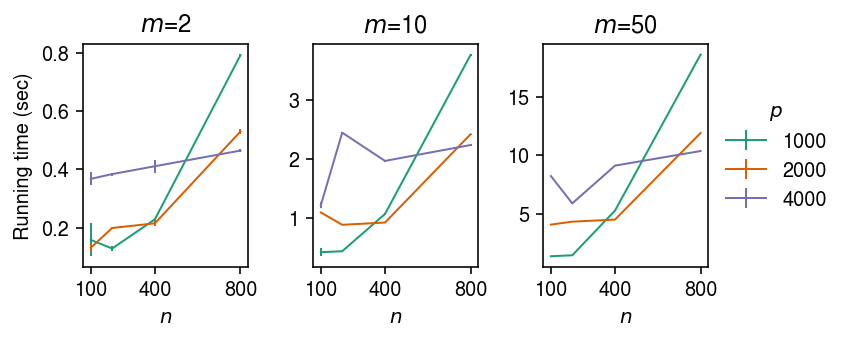
Benchmark running time for a single choice of \(\lambda_1, \lambda_2\) from cold initialization \(\mb = \boldsymbol{0}\).
def evaluate_running_time(n, p, m): f = gtexpred.lasso.sparse_multi_task_lasso X, Y, B = simulate_identical_effects(n=n, p=p, m=m, n_causal=1, pve=0.1, seed=0) res = timeit.repeat(lambda: f(X, Y, lambda1=1, lambda2=1), repeat=10, number=1, globals=locals()) return np.array(res).mean(), np.array(res).std() result = [] for n in (100, 200, 400, 800): for p in (1000, 2000, 4000): for m in (2, 10, 50): mean, std = evaluate_running_time(n, p, m) result.append((n, p, m, mean, std)) result = pd.DataFrame(result) result.columns = ['n', 'p', 'm', 'mean', 'std'] result.to_csv('/project2/mstephens/aksarkar/projects/gtex-pred/data/sparse-multi-task-lasso-runtime.txt.gz', sep='\t', compression='gzip')
sbatch --partition=broadwl -n1 -c1 --exclusive --time=60:00 #!/bin/bash source activate fqtl python <<EOF <<imports>> <<simulation>> <<evaluate-running-time>> EOF
Read the results.
result = pd.read_csv('/project2/mstephens/aksarkar/projects/gtex-pred/data/sparse-multi-task-lasso-runtime.txt.gz', sep='\t', index_col=0)
Plot run time as a function of \(n, m, p\). Error bars denote 2 standard deviations over 10 trials.
cm = plt.get_cmap('Dark2') plt.clf() fig, ax = plt.subplots(1, 3) fig.set_size_inches(6, 2.5) for a, (m, g) in zip(ax, result.groupby('m')): for i, (p, g2) in enumerate(g.groupby('p')): a.errorbar(g2['n'], g2['mean'], yerr=2 * g2['std'], c=cm(i), label=p, lw=1) a.set_xticks([100, 400, 800]) a.set_xlabel('$n$') a.set_title(f'$m$={m}') ax[0].set_ylabel('Running time (sec)') ax[-1].legend(title='$p$', frameon=False, loc='center left', bbox_to_anchor=(1, .5)) fig.tight_layout()
Lasso path
The coordinate descent update (assuming bjk ≥ 0) is
\[ b_{jk} := \min\left(\frac{\mr_{\cdot k} \mx_{\cdot j} - \lambda_1}{(\mx' \mx)_{jj} + \lambda_2 / \Vert \mb_{j \cdot} \Vert_2}, 0\right) \]
As expected, we recover the lasso update if \(\lambda_2 = 0\). For the lasso, the update reveals that any \(b_{jk}\) for which \(\mr_{\cdot k} \mx_{\cdot j} < \lambda_1\) will be shrunk to 0. This gives a way to find the maximum \(\lambda_1\) for a given data set:
\[ \lambda_{1,\max} = \max \vert \mx' \my / n \vert \]
def est_sparse_multi_task_lasso_path(X, Y, lambda2, trace=0, **kwargs): print(f'estimating path for lambda2 = {lambda2}') xty = X.T @ Y lam_max = np.abs(xty).max() grid = np.geomspace(lam_max, .01, 50) Bhat = xty / X.shape[0] path = dict() for a in grid: Bhat, _ = mtlasso.lasso.sparse_multi_task_lasso(X, Y, init=Bhat, lambda1=a, lambda2=lambda2, **kwargs) path[a] = Bhat[trace].copy() return pd.DataFrame.from_dict(path)
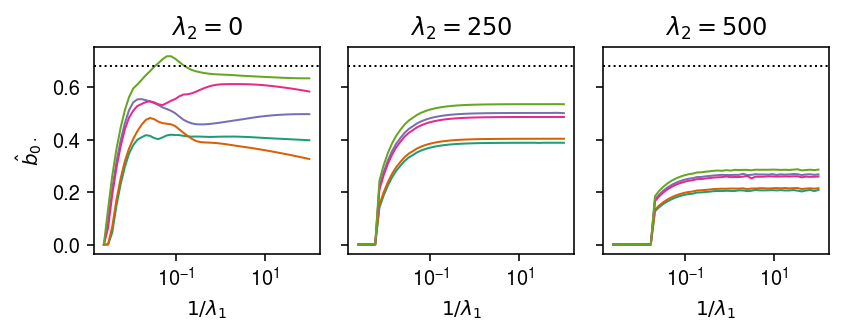
Simulate data for 5 conditions and a single causal variant which has identical effect in all conditions. Look at the paths of the coefficients for the simulated causal variant.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=1, pve=0.1, seed=1) paths = { lambda2: est_sparse_multi_task_lasso_path(X, Y, lambda2=lambda2, atol=1e-4, max_iter=500) for lambda2 in (0, 250, 500) }
cm = plt.get_cmap('Dark2') plt.clf() fig, ax = plt.subplots(1, 3, sharey=True) fig.set_size_inches(6, 2.5) for a, (lambda2, path) in zip(ax, paths.items()): a.set_xscale('log') for k, row in path.iterrows(): a.plot(1 / path.columns, row, lw=1, c=cm(k)) a.axhline(y=B[0,0], ls=':', c='k', lw=1) a.set_xlabel('$1 / \lambda_1$') a.set_title(f'$\lambda_2 = {lambda2}$') ax[0].set_ylabel('$\hat{b}_{0\cdot}$') fig.tight_layout()
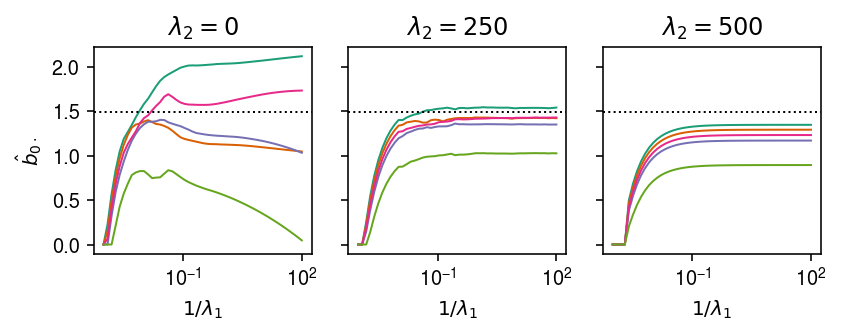
Simulate multiple causal variants.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=3, pve=0.1, seed=0) paths = { lambda2: est_sparse_multi_task_lasso_path(X, Y, lambda2=lambda2, atol=1e-4, max_iter=500) for lambda2 in (0, 250, 500) }
cm = plt.get_cmap('Dark2') plt.clf() fig, ax = plt.subplots(1, 3, sharey=True) fig.set_size_inches(6, 2.5) for a, (lambda2, path) in zip(ax, paths.items()): a.set_xscale('log') for k, row in path.iterrows(): a.plot(1 / path.columns, row, lw=1, c=cm(k)) a.axhline(y=B[0,0], ls=':', c='k', lw=1) a.set_xlabel('$1 / \lambda_1$') a.set_title(f'$\lambda_2 = {lambda2}$') ax[0].set_ylabel('$\hat{b}_{0\cdot}$') fig.tight_layout()
Start from the oracle initialization to understand why the estimated coefficients are not driven to be equal.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=3, pve=0.1, seed=0) Bhat, Bhat0 = mtlasso.lasso.sparse_multi_task_lasso(X, Y, init=B.copy(), lambda1=0, lambda2=200, atol=1e-4) Bhat[:3]
array([[ 1.57120005, 1.42403555, 1.36905244, 1.49823112, 1.02940123], [-0.1172862 , -0.11772508, -0.14990959, -0.12432973, -0.00778592], [ 1.51250475, 1.25522477, 0.89798756, 1.04866882, 1.42675512]])
Idealized simulation
Simulate an easy example, and verify the method worked.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=10, n_causal=1, pve=0.1, seed=0) # Hold out validation data to evaluate the final model Xt, Xv, Yt, Yv = skms.train_test_split(X, Y, test_size=0.1) mx = Xt.mean(axis=0) my = Yt.mean(axis=0)
Fix \(\lambda_1, \lambda_2\).
Bhat = gtexpred.lasso.sparse_multi_task_lasso(Xt - mx, Yt - my, lambda1=150, lambda2=100, atol=1e-8)
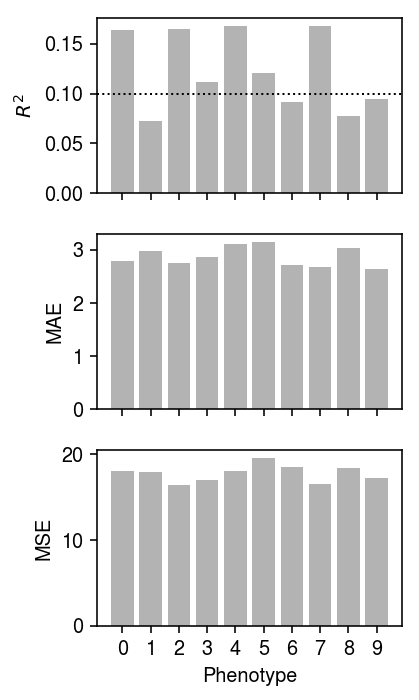
Make sure the model worked on the training data.
plt.clf() fig, ax = plt.subplots(3, 1, sharex=True) fig.set_size_inches(3, 5) Yhat = (Xt - mx).dot(Bhat) ax[0].bar(np.arange(10), skm.r2_score(Yt - my, Yhat, multioutput='raw_values'), color='0.7') ax[0].axhline(y=0.1, ls=':', lw=1, c='k') ax[0].set_ylabel('$R^2$') ax[1].bar(np.arange(10), np.median(abs(Yt - my - Yhat), axis=0), color='0.7') ax[1].set_ylabel('MAE') ax[2].bar(np.arange(10), skm.mean_squared_error(Yt - my, Yhat, multioutput='raw_values'), color='0.7') ax[2].set_ylabel('MSE') ax[2].set_xlabel('Phenotype') for a in ax: a.set_xticks(np.arange(10)) ax[2].set_xticklabels(np.arange(10)) plt.tight_layout()
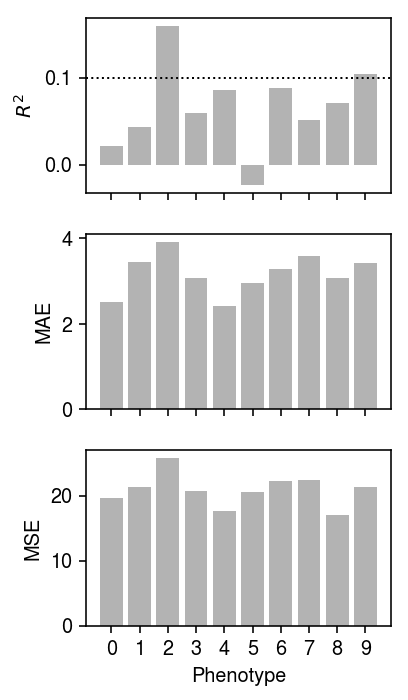
Evaluate the model on the validation data.
plt.clf() fig, ax = plt.subplots(3, 1, sharex=True) fig.set_size_inches(3, 5) Yhat = (Xv - mx).dot(Bhat) ax[0].bar(np.arange(10), skm.r2_score(Yv - my, Yhat, multioutput='raw_values'), color='0.7') ax[0].axhline(y=0.1, ls=':', lw=1, c='k') ax[0].set_ylabel('$R^2$') ax[1].bar(np.arange(10), np.median(abs(Yv - my - Yhat), axis=0), color='0.7') ax[1].set_ylabel('MAE') ax[2].bar(np.arange(10), skm.mean_squared_error(Yv - my, Yhat, multioutput='raw_values'), color='0.7') ax[2].set_ylabel('MSE') ax[2].set_xlabel('Phenotype') for a in ax: a.set_xticks(np.arange(10)) ax[2].set_xticklabels(np.arange(10)) plt.tight_layout()
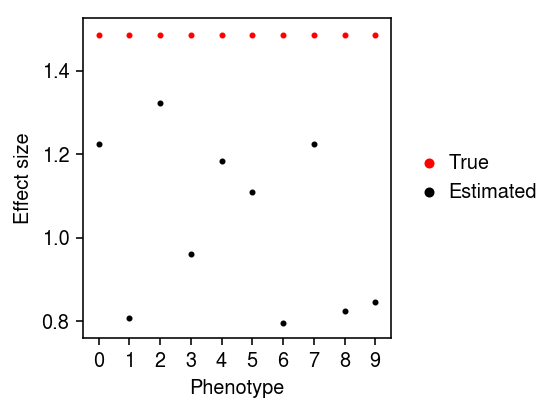
Compare the fitted coefficients at the true causal variant to the true coefficients.
plt.clf() plt.gcf().set_size_inches(4, 3) plt.scatter(np.arange(10), B[0,:], color='r', label='True', s=4) plt.scatter(np.arange(10), Bhat[0,:], color='k', label='Estimated', s=4) plt.legend(frameon=False, loc='center left', bbox_to_anchor=(1, .5), handletextpad=0, markerscale=2) plt.xticks(np.arange(10), np.arange(10)) plt.xlabel('Phenotype') plt.ylabel('Effect size') plt.tight_layout()
Cross-validation
Simulate an easy example.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=1, pve=0.1, seed=0) # Hold out validation data to evaluate the final model Xt, Xv, Yt, Yv = skms.train_test_split(X, Y, test_size=0.1) mx = Xt.mean(axis=0) my = Yt.mean(axis=0)
Tune \(\lambda_1, \lambda_2\) by 5-fold cross validation within the training data.
# c.f. https://github.com/scikit-learn/scikit-learn/blob/1495f69242646d239d89a5713982946b8ffcf9d9/sklearn/linear_model/coordinate_descent.py#L112 grid = np.geomspace(.1, 1, 10) * Xt.shape[0] cv_scores = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Yt, cv=skms.KFold(n_splits=5), lambda1=grid, lambda2=grid, max_iter=500, verbose=True) cv_scores = pd.DataFrame(cv_scores) cv_scores.columns = ['fold', 'lambda1', 'lambda2', 'mse']
cv_scores.to_csv('/scratch/midway2/aksarkar/txpred/full-data-cv-scores.txt.gz', sep='\t')
cv_scores = pd.read_csv('/scratch/midway2/aksarkar/txpred/full-data-cv-scores.txt.gz', sep='\t')
lambda1, lambda2 = cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin() lambda1, lambda2
(45.0, 208.87149751257505)
Look at the best performing penalty weight choices.
cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).sort_values().head().reset_index()
lambda1 lambda2 mse 0 45.000000 208.871498 18.675936 1 75.064524 161.721615 18.697633 2 58.119735 208.871498 18.705495 3 58.119735 161.721615 18.710209 4 45.000000 269.767913 18.737840
Fit the model on the full training data using the best \(\lambda_1, \lambda_2\).
Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1=lambda1, lambda2=lambda2)
Score the final model on the validation data.
pd.DataFrame({
'r2': skm.r2_score(Yv - my - B0hat, (Xv - mx).dot(Bhat), multioutput='raw_values'),
'mse': np.square(Yv - my - B0hat - (Xv - mx).dot(Bhat)).mean(axis=0),
'mae': np.median(np.abs(Yv - my - B0hat - (Xv - mx).dot(Bhat)), axis=0)
})
r2 mse mae 0 0.212076 23.835363 3.408341 1 0.074575 22.467354 3.564529 2 0.060348 19.873910 2.961368 3 0.116312 22.517498 3.416149 4 0.089665 19.693309 3.128175
Try fixing \(\lambda_1 = 0\)
cv_scores_lam1_0 = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Yt, cv=skms.KFold(n_splits=5), lambda1=[0.], lambda2=grid, max_iter=500, verbose=True) cv_scores_lam1_0 = pd.DataFrame(cv_scores_lam1_0) cv_scores_lam1_0.columns = ['fold', 'lambda1', 'lambda2', 'mse']
cv_scores_lam1_0.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).sort_values().head().reset_index()
lambda1 lambda2 mse 0 0.0 300.150811 18.726872 1 0.0 372.063809 18.738914 2 0.0 242.137255 18.856089 3 0.0 195.336637 19.084469 4 0.0 157.581707 19.512256
lambda1, lambda2 = cv_scores_lam1_0.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin() Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1=lambda1, lambda2=lambda2) pd.DataFrame({ 'r2': skm.r2_score(Yv - my - B0hat, (Xv - mx).dot(Bhat), multioutput='raw_values'), 'mse': np.square(Yv - my - B0hat - (Xv - mx).dot(Bhat)).mean(axis=0), 'mae': np.median(np.abs(Yv - my - B0hat - (Xv - mx).dot(Bhat)), axis=0) })
r2 mse mae 0 0.208358 23.947845 3.363572 1 0.075627 22.441819 3.630488 2 0.063343 19.810545 2.960831 3 0.123200 22.341967 3.392123 4 0.088933 19.709142 3.070840
SMBO vs. CV
The goal of CV is to find the parameters of the training algorithm (here, \(\lambda_1\) and \(\lambda_2\)) that minimizes a loss (here, mean-squared error). Above, we evaluated \(k\)-fold CV, which takes \(k \times n^2\) model fits, where \(n\) is the size of the grid of candidate values for \(\lambda_1\) and \(\lambda_2\) This approach scales poorly for high resolution grids (large \(n\)), and self-evidently spends a lot of time on values of the parameters that are highly unlikely to be optimal.
One could in principle do better using sequential model-based optimization (SMBO; e.g., Bergstra et al. 2011). This approach treats the loss as an unknown function \(f\) (which is expensive to evaluate) to be modeled; it uses each evaluation of \(f\) to update a model of \(f\); and it uses the current model of \(f\) to pick the next point at which to evaluate \(f\). Bergstra et al. 2011 propose the Tree of Parzen Estimators as a model of \(f\). In principle, this approach should be able to more efficiently search the space, by focusing effort on parameter values near the optimum.
Simulate the same example as above.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=1, pve=0.1, seed=0) # Hold out validation data to evaluate the final model Xt, Xv, Yt, Yv = skms.train_test_split(X, Y, test_size=0.1) mx = Xt.mean(axis=0) my = Yt.mean(axis=0)
Specify CV loss as a black-box function.
Remark One could in principle not do full \(k\)-fold CV within this function; however, then the SMBO algorithm needs to model noisy function evaluations, which seems to be an open problem.
def loss(theta, X, Y, **kwargs): lambda1, lambda2 = theta scores = [] init = np.zeros((X.shape[1], Y.shape[1])) for train_idx, test_idx in skms.KFold(n_splits=5).split(X, Y): Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(X[train_idx], Y[train_idx], lambda1, lambda2, init=init, **kwargs) score = mtlasso.lasso._mse(X[test_idx], Y[test_idx] - B0hat, Bhat) init = Bhat scores.append(score) return {'loss': np.array(scores).mean(), 'status': hp.STATUS_OK}
Run SMBO.
np.random.seed(1) trials = hp.Trials() opt = hp.fmin( ft.partial(loss, X=Xt, Y=Yt, max_iter=500), space=[hp.hp.loguniform('lambda1', np.log(.1 * Xt.shape[0]), np.log(Xt.shape[0])), hp.hp.loguniform('lambda2', np.log(.1 * Xt.shape[0]), np.log(Xt.shape[0]))], algo=hp.tpe.suggest, trials=trials, max_evals=20)
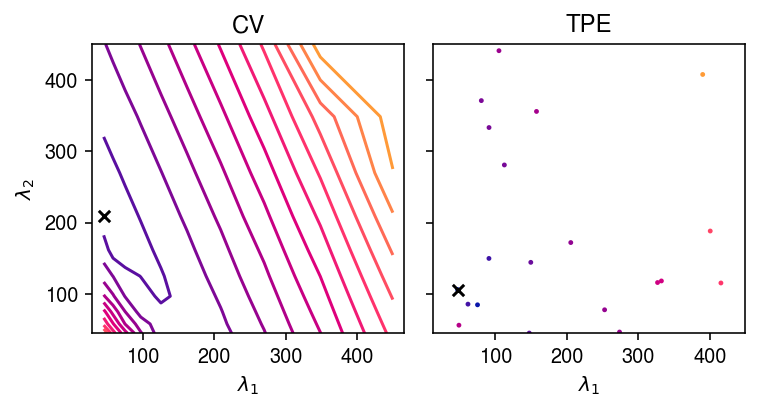
Look at the contours of the cross-validation loss as a function of \(\lambda_1, \lambda_2\), based on the grid of evaluations in full CV, and compare against the candidate points evaluated in SMBO.
cm = colorcet.cm['bmy'] norm = matplotlib.colors.Normalize(vmin=18., vmax=22.) plt.clf() fig, ax = plt.subplots(1, 2, sharey=True) fig.set_size_inches(5.5, 3) for a in ax: a.set_aspect('equal', adjustable='datalim') query = cv_scores.pivot_table(index='lambda2', columns='lambda1', values='mse').values ax[0].contour(grid, grid, query, cmap=cm, norm=norm, levels=15) ax[0].scatter(lambda1, lambda2, s=32, marker='x', c='k') ax[0].set_xlabel('$\lambda_1$') ax[0].set_ylabel('$\lambda_2$') ax[0].set_title('CV') query = pd.DataFrame([{k: v[0] for k, v in x['misc']['vals'].items()} for x in trials.trials]) query['loss'] = [x['result']['loss'] for x in trials.trials] ax[1].scatter(query['lambda1'], query['lambda2'], c=cm(norm(query['loss'])), s=2) ax[1].scatter(opt['lambda1'], opt['lambda2'], s=32, marker='x', c='k') ax[1].set_xlabel('$\lambda_1$') ax[1].set_title('TPE') fig.tight_layout()
Report the optimal \(\lambda_1, \lambda_2\) estimated by SMBO.
opt
{'lambda1': 46.93776994095178, 'lambda2': 106.27197065892976}
Fit sparse multi-task lasso with that choice of \(\lambda_1, \lambda_2\), and evaluate the performance on the validation data.
Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1=opt['lambda1'], lambda2=opt['lambda2']) pd.DataFrame({ 'r2': skm.r2_score(Yv - my - B0hat, (Xv - mx).dot(Bhat), multioutput='raw_values'), 'mse': np.square(Yv - my - B0hat - (Xv - mx).dot(Bhat)).mean(axis=0), 'mae': np.median(np.abs(Yv - my - B0hat - (Xv - mx).dot(Bhat)), axis=0) })
r2 mse mae 0 0.192476 24.428296 3.626432 1 0.024523 23.682503 3.755742 2 0.076187 19.538899 3.059620 3 0.039539 24.473773 3.719337 4 0.019015 21.221685 2.753583
This methodology does slightly worse than full CV, but runs in a fraction of the wall clock time.
Missing data
Simulate an easy example.
X, Y, B = simulate_identical_effects(n=500, p=1000, m=5, n_causal=1, pve=0.1, seed=0) # Hold out validation data to evaluate the final model Xt, Xv, Yt, Yv = skms.train_test_split(X, Y, test_size=0.1) mx = Xt.mean(axis=0) my = Yt.mean(axis=0)
Suppose not every phenotype is observed in the training data. As an example, mask 50% of the training phenotype matrix entries.
Z = np.random.uniform(size=Yt.shape) < 0.5 Ytm = np.ma.masked_array(Yt, mask=Z)
Fit the model on the observed training data.
grid = np.geomspace(.1, 1, 10) * Xt.shape[0] cv_scores = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Ytm, cv=skms.KFold(n_splits=5), lambda1=grid, lambda2=grid, max_iter=200, verbose=True) cv_scores = pd.DataFrame(cv_scores) cv_scores.columns = ['fold', 'lambda1', 'lambda2', 'mse'] lambda1, lambda2 = cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin() Bhat0 = mtlasso.lasso.sparse_multi_task_lasso(Xt, Ytm, lambda1, lambda2)
cv_scores.to_csv('/scratch/midway2/aksarkar/txpred/masked-data-cv-scores.txt.gz', sep='\t')
For comparison, fit the model on the full training data.
cv_scores = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Yt, cv=skms.KFold(n_splits=5), lambda1=grid, lambda2=grid, max_iter=200) cv_scores = pd.DataFrame(cv_scores) cv_scores.columns = ['fold', 'lambda1', 'lambda2', 'mse'] lambda1, lambda2 = cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin() Bhat1 = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1, lambda2)
Compare the fitted values of the two models.
plt.clf() fig, ax = plt.subplots(1, 5, sharey=True) fig.set_size_inches(7.5, 2.5) for i, a in enumerate(ax): a.set_aspect('equal', adjustable='datalim') a.scatter(Xv @ Bhat0[0][:,i] + Bhat0[1][:,i], Xv @ Bhat1[0][:,i] + Bhat1[1][:,i], s=1, c='k') a.set_title(f'Phenotype {i}') ax[0].set_ylabel('Full data out-of-sample\npredicted value') a = fig.add_subplot(111, xticks=[], yticks=[], frameon=False) a.set_xlabel('Masked data out-of-sample predicted value', labelpad=20) fig.tight_layout()
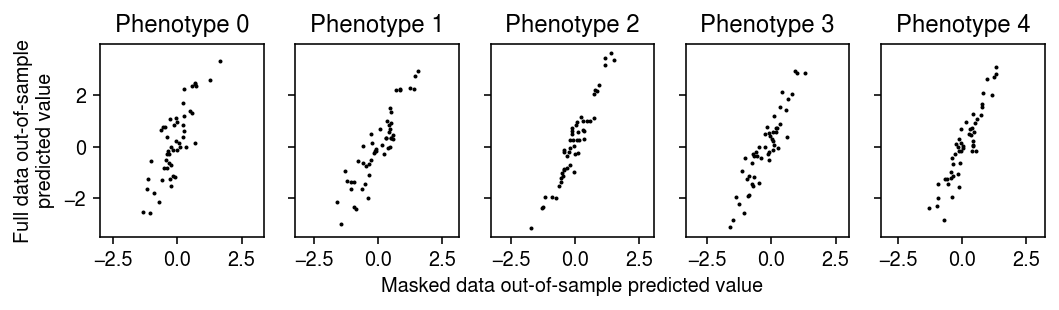
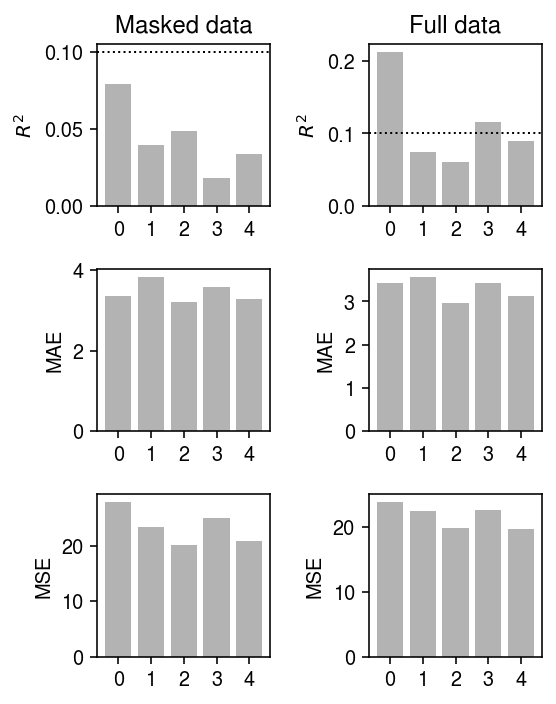
Evaluate the fits on the validation data.
plt.clf() fig, ax = plt.subplots(3, 2) fig.set_size_inches(4, 5) for a, b, t in zip(ax.T, (Bhat0, Bhat1), ('Masked data', 'Full data')): Yhat = Xv @ b[0] + b[1] a[0].bar(np.arange(5), skm.r2_score(Yv - my, Yhat, multioutput='raw_values'), color='0.7') a[0].axhline(y=0.1, ls=':', lw=1, c='k') a[0].set_ylabel('$R^2$') a[1].bar(np.arange(5), np.median(abs(Yv - my - Yhat), axis=0), color='0.7') a[1].set_ylabel('MAE') a[2].bar(np.arange(5), skm.mean_squared_error(Yv - my, Yhat, multioutput='raw_values'), color='0.7') a[2].set_ylabel('MSE') a[0].set_title(t) for a in ax.ravel(): a.set_xticks(np.arange(5)) fig.tight_layout()
Estimate the difference in the estimated effect sizes.
np.linalg.norm(Bhat0[0] - Bhat1[0])
1.8253611488999593
Comparison to univariate elastic-net
Simulate an easy case.
rng = np.random.default_rng(1) n = 1000 p = 5000 m = 10 X = rng.normal(size=(n, p)) B = np.zeros((p, m)) B[0,:3] = rng.normal(size=(1, 3)) B[1,3:] = rng.normal(size=(1, 7)) Y = X @ B + rng.normal(size=(n, m))
Split the simulated data into training and validation.
Xt, Xv, Yt, Yv = skms.train_test_split(X, Y, test_size=0.1, random_state=1)
Fit univariate elastic-net models for each simulated response, using 4-fold CV on the penalty weights.
enets = [] for k in range(Y.shape[1]): print(f'fitting {k}') fit = sklm.ElasticNetCV(cv=4, n_jobs=8, normalize=True, fit_intercept=True).fit(Xt, Yt[:,k]) enets.append(fit)
Report the training and validation MSE for each response.
pd.DataFrame({
'train': {k: skm.mean_squared_error(Xt @ enets[k].coef_ + enets[k].intercept_, Yt[:,k]) for k in range(10)},
'val': {k: skm.mean_squared_error(Xv @ enets[k].coef_ + enets[k].intercept_, Yv[:,k]) for k in range(10)},
})
train val 0 1.162114 0.736277 1 1.098416 1.092427 2 0.790721 2.129253 3 1.010724 1.366760 4 1.303171 1.805048 5 1.023614 1.151061 6 1.075434 0.959652 7 0.764977 3.814086 8 1.025173 1.657425 9 1.096054 1.210242
Fit sparse multi-task lasso, using 4-fold CV to select the optimal lambda2, and taking the optimal lambda1 on the path for that choice.
grid = np.geomspace(.1, 1, 10) * Xt.shape[0] cv_scores = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Yt, cv=skms.KFold(n_splits=4), lambda1=grid, lambda2=grid, max_iter=500, verbose=True) cv_scores = pd.DataFrame(cv_scores) cv_scores.columns = ['fold', 'lambda1', 'lambda2', 'mse'] lambda1, lambda2 = cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin() Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1=lambda1, lambda2=lambda2)
Report the training and validation MSE for each response.
pd.DataFrame({
'train': {k: skm.mean_squared_error((Xt @ Bhat + B0hat)[:,k], Yt[:,k]) for k in range(10)},
'val': {k: skm.mean_squared_error((Xv @ Bhat + B0hat)[:,k], Yv[:,k]) for k in range(10)},
})
train val 0 1.089945 0.711197 1 1.007758 0.990568 2 1.032572 0.795654 3 0.998397 1.069531 4 1.009947 1.078685 5 1.013060 1.142606 6 1.054464 0.933449 7 1.057387 0.965998 8 1.021355 1.004041 9 1.011801 1.048657
Plot the simulated effect sizes and MSE for the fitted models.
plt.clf() fig, ax = plt.subplots(3, 1, sharex=True) fig.set_size_inches(4, 5.5) i = ax[0].imshow(B[:4], cmap=colorcet.cm['cwr'], vmin=-3, vmax=3) ax[0].set_aspect('auto') fig.colorbar(i, label='Ground truth effect size', ax=ax[0], shrink=0.5, orientation='horizontal') ax[1].bar(range(10), [skm.mean_squared_error(Xt @ enets[k].coef_ + enets[k].intercept_, Yt[:,k]) for k in range(10)], color='k', alpha=0.5, label='Training') ax[1].bar(range(10), [skm.mean_squared_error(Xv @ enets[k].coef_ + enets[k].intercept_, Yv[:,k]) for k in range(10)], color='r', alpha=0.5, label='Validation') ax[2].bar(range(10), [skm.mean_squared_error((Xt @ Bhat + B0hat)[:,k], Yt[:,k]) for k in range(10)], color='k', alpha=0.5, label='Training') ax[2].bar(range(10), [skm.mean_squared_error((Xv @ Bhat + B0hat)[:,k], Yv[:,k]) for k in range(10)], color='r', alpha=0.5, label='Validation') ax[0].set_ylabel('Predictor') ax[1].set_ylabel('Elastic-net MSE') ax[2].set_ylabel('mtlasso MSE') ax[-1].set_xlabel('Response') for a in ax: a.set_xticks(range(10)) for a in ax[1:]: a.set_ylim(0, 4) a.legend(frameon=False, loc='center left', bbox_to_anchor=(1, .5)) fig.tight_layout()
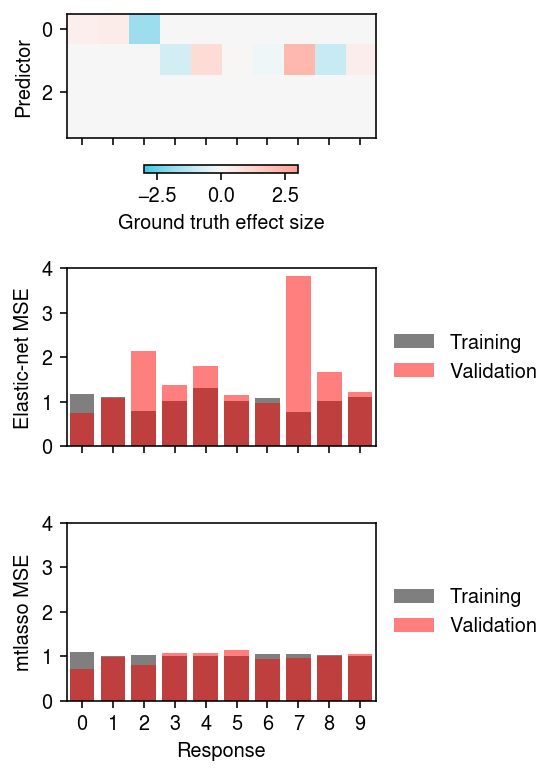
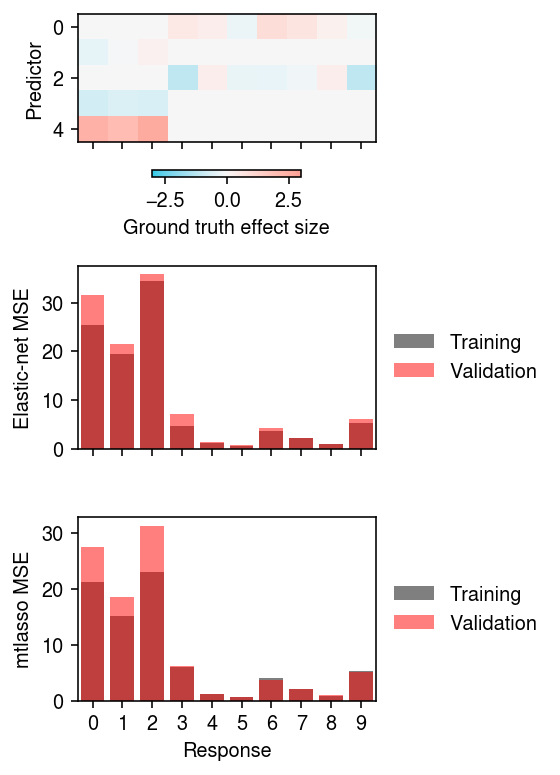
Look at Fabio's example.
dat = rpy2.robjects.r['readRDS']('/project2/mstephens/fmorgante/test.rds')
X, Y, B, V, _, _, B0, _, idx, _ = [np.array(t) for t in dat]
n = 900 Xt = X[:n] Xv = X[n:] Yt = Y[:n] Yv = Y[n:]
Fit univariate elastic nets.
enets = [] for k in range(Y.shape[1]): print(f'fitting {k}') fit = sklm.ElasticNetCV(cv=4, n_jobs=8, normalize=True, fit_intercept=True).fit(Xt, Yt[:,k]) enets.append(fit)
Report the training and validation MSE for each response.
pd.DataFrame({
'train': {k: skm.mean_squared_error(Xt @ enets[k].coef_ + enets[k].intercept_, Yt[:,k]) for k in range(10)},
'val': {k: skm.mean_squared_error(Xv @ enets[k].coef_ + enets[k].intercept_, Yv[:,k]) for k in range(10)},
})
train val 0 25.362915 31.492174 1 19.354158 21.605593 2 34.449236 35.804181 3 4.740143 7.053288 4 1.282722 1.391711 5 0.662963 0.735404 6 3.736664 4.286861 7 2.128571 2.280445 8 0.939318 1.009634 9 5.289383 6.038868
Fit sparse multi-task lasso, using 4-fold CV to select the optimal lambda2, and taking the optimal lambda1 on the path for that choice.
grid = np.geomspace(.1, 1, 10) * Xt.shape[0] cv_scores = mtlasso.lasso.sparse_multi_task_lasso_cv( Xt, Yt, cv=skms.KFold(n_splits=4), lambda1=grid, lambda2=grid, max_iter=500, verbose=True) lambda1, lambda2 = cv_scores.groupby(['lambda1', 'lambda2'])['mse'].agg(np.mean).idxmin()
Bhat, B0hat = mtlasso.lasso.sparse_multi_task_lasso(Xt, Yt, lambda1=lambda1, lambda2=lambda2)
np.savez('/scratch/midway2/aksarkar/txpred/fabio-ex.npz', Bhat=Bhat, B0hat=B0hat)
Report the training and validation MSE for each response.
pd.DataFrame({
'train': {k: skm.mean_squared_error((Xt @ Bhat + B0hat)[:,k], Yt[:,k]) for k in range(10)},
'val': {k: skm.mean_squared_error((Xv @ Bhat + B0hat)[:,k], Yv[:,k]) for k in range(10)},
})
train val 0 21.119216 27.357582 1 15.093317 18.467278 2 22.991694 31.168670 3 5.986364 6.094789 4 1.161488 1.240754 5 0.640629 0.675358 6 4.054401 3.756979 7 2.015473 2.042536 8 0.869960 0.919764 9 5.217864 5.155873
plt.clf() fig, ax = plt.subplots(3, 1, sharex=True) fig.set_size_inches(4, 5.5) i = ax[0].imshow(B[idx - 1], cmap=colorcet.cm['cwr'], vmin=-3, vmax=3) ax[0].set_aspect('auto') fig.colorbar(i, label='Ground truth effect size', ax=ax[0], shrink=0.5, orientation='horizontal') ax[1].bar(range(10), [skm.mean_squared_error(Xt @ enets[k].coef_ + enets[k].intercept_, Yt[:,k]) for k in range(10)], color='k', alpha=0.5, label='Training') ax[1].bar(range(10), [skm.mean_squared_error(Xv @ enets[k].coef_ + enets[k].intercept_, Yv[:,k]) for k in range(10)], color='r', alpha=0.5, label='Validation') ax[2].bar(range(10), [skm.mean_squared_error((Xt @ Bhat + B0hat)[:,k], Yt[:,k]) for k in range(10)], color='k', alpha=0.5, label='Training') ax[2].bar(range(10), [skm.mean_squared_error((Xv @ Bhat + B0hat)[:,k], Yv[:,k]) for k in range(10)], color='r', alpha=0.5, label='Validation') ax[0].set_ylabel('Predictor') ax[1].set_ylabel('Elastic-net MSE') ax[2].set_ylabel('mtlasso MSE') ax[-1].set_xlabel('Response') for a in ax: a.set_xticks(range(10)) for a in ax[1:]: a.legend(frameon=False, loc='center left', bbox_to_anchor=(1, .5)) fig.tight_layout()