Tips-and-Tricks¶
This workshop is designed to highlight some of the lesser known features and useful concepts for getting the most out of RCC’s Midway cluster.
Accessing Midway¶
Mosh¶
Mosh is an MIT-developed replacement for SSH that can tolerate roaming and intermittent
connectivity. It is convenient when using a laptop and moving from place to place.
Install it on your local machine, and then add the following line to your ~/.bashrc file
on Midway (or ~/.zshenv if you use zsh):
module load mosh
Then, connect to Midway with
mosh midway.rcc.uchicago.edu
Graphical environment¶
It can be convenient to use Midway with a desktop environment; some software packages like Rstudio or Mathematica have their own GUIs. You can connect with ssh -X midway.rcc.uchicago.edu, but this can be very slow, particularly when off-campus.
RCC recommends Connecting with ThinLinc for graphical connections. There’s a desktop client you can download and use, or, for the easiest way to access Midway with a GUI, go to our web interface at http://midway.rcc.uchicago.edu.
Software¶
Modules¶
Midway is a general-purpose computing system available to a large number of users in a variety of fields. To support these users, RCC provides a large number of libraries and applications that have been compiled and tuned for Midway. To maintain a consistent user environment, the- RCC uses a modules system for configuring a user’s environment.
Basic module commands:
| Command | Description |
|---|---|
| module avail [name] | lists modules matching [name] (all if ‘name’ empty) |
| module load [name] | loads the named module |
| module unload [name] | unloads the named module |
| module list | lists the modules currently loaded for the user |
| module display name | shows the effects of loading module name |
Example:
$ module display python/2.7-2015q2
-------------------------------------------------------------------
/software/modulefiles/python/2.7-2015q2:
module-whatis setup python 2.7-2015q2 compiled with the system compiler
conflict python
module load mkl/11.2 texlive/2012 hdf5/1.8 netcdf/4.2 graphviz/2.28 qt/4.8 geos/3.4 gdal/1.11 postgresql/9.2
prepend-path PATH /software/python-2.7-2015q2-el6-x86_64/bin
prepend-path LD_LIBRARY_PATH /software/python-2.7-2015q2-el6-x86_64/lib
prepend-path LIBRARY_PATH /software/python-2.7-2015q2-el6-x86_64/lib
prepend-path PKG_CONFIG_PATH /software/python-2.7-2015q2-el6-x86_64/lib/pkgconfig
prepend-path CPATH /software/python-2.7-2015q2-el6-x86_64/include
prepend-path MANPATH /software/python-2.7-2015q2-el6-x86_64/share/man
prepend-path MANPATH /software/python-2.7-2015q2-el6-x86_64/man
prepend-path QT_PLUGIN_PATH /software/python-2.7-2015q2-el6-x86_64/share/qt_plugins
setenv SCONS_LIB_DIR /software/python-2.7-2015q2-el6-x86_64/lib/python2.7/site-packages/scons-2.3.4-py2.7.egg/scons-2.3.4/
setenv NLTK_DATA /project/databases/nltk_data
-----------------------------------------------
This command shows that the python/2.7-2015q2 module loads a number of dependent modules (mkl, hdf5, qt, etc),
and sets a number of environment variables necesary to locate include and library files, as well as affecting program execution.
Custom Modules¶
Users and research groups can define their own modules which can simplify package management. First create a module file
and use commands like load, conflict, and setenv to modify your environment.
#%Module1.0#####################################################################
module load WMAP/9
module load Planck/1.0-2
module load intelmpi/4.1+intel-13.1 mkl/10.3 cfitsio/3
conflict "CosmoMC"
setenv PLANCKLIKE cliklike
setenv CLIKPATH \$PLC_DIR
The module command searches your “module path” to locate these files. By default, only the system-wide directory
/software/modules is in your search path. Use the command module use to add your own directory, or use the
convenient RCC provided module use.own to automatically add $HOME/private_modules.
$ module use /project/kicp/opt/modules
$ module avail
------------------------------- /project/kicp/opt/modules/ -------------------------------------------------------
WMAP/7 WMAP/9+intel-15.0 Planck/1.0+intel-14.0 Planck/1.0-2 CosmoMC/May14
WMAP/9(default) Planck/1.0 Planck/1.0+intel-15.0 CosmoMC/CosmoMC(default)
Installing software locally¶
RCC is very good about installing software as a module, however some software may not be appropriate. Examples are software that requires user-level customization, or a rapid release schedule. Users can always install software into their home directory.
Compiled programs and libraries in UNIX often use autoconf based builds, which provide an executable configure
script. These programs often by default install into a system-wide location, such as /usr/local. Instead, use
the --prefix option to install into your home or project directory.
./configure --prefix=$HOME
Packages for other specialized environments such as R or python can be installed using specific commands,
which we will discuss in later sections.
Running Jobs¶
sinteractive¶
In general, the login nodes can be used for development and to run short, resource-light programs, and scripts can
be submitted to the cluster with sbatch. For interactive computing – programming with short development cycles,
or with a R/Python/Matlab REPL – use sinteractive. sinteractive can be used with the same flags as sbatch, for example:
sinteractive --ntasks-per-node=4 --mem-per-cpu=8000
Slurm Environment Variables¶
Slurm sets a number of environment variables that can be queried within a job script to get information about the environment, for example:
| SLURM_JOB_ID | The ID of the job allocation |
| SLURM_JOB_NAME | Name of the job |
| SLURM_JOB_NODELIST | List of nodes allocated to the job |
| SLURM_JOB_NUM_NODES | Number of nodes in the job’s allocation. |
| SLURM_NTASKS_PER_NODE | Number of tasks initiated on each node. |
| SLURM_NTASKS | Total number of tasks (usually, CPUs) allocated. |
A full list is available in Slurm’s documentation here: https://computing.llnl.gov/linux/slurm/sbatch.html
For example, the following Python script queries the number of CPU allocated to it:
#!/usr/bin/env python
import os
print "I am using %s CPUs" % os.environ['SLURM_NTASKS']
In reality, a parallelized Python script might use the number of CPUs allocated to it in order to configure the distribution of data to different processes.
Array jobs¶
The simplest way to use parallelism is often to run the same program on a large number of different inputs. This is often called SIMD parallelism (single instruction, multiple data). Slurm provides array jobs which can simplify this style of parallelization. A single script is submitted multiple times, with a different setting of the environment variable SLURM_ARRAY_TASK_ID. When files are named cleverly, this can make it easy to specify a unique input for each instance of the script. Each subjob is scheduled indepedently, which may reduce the total time jobs are waiting in the queue.
For example, say there is a folder with 100 JPEG images, named 1.jpg, 2.jpg, ..., 100.jpg. The following script will be run 100 times, resizing and converting all the images using the ImageMagick utility:
#!/bin/bash
#SBATCH --job-name=convert
#SBATCH --output=convert_%A_%a.out
#SBATCH --array=1-100
convert $SLURM_ARRAY_TASK_ID.jpg -resize 50% $SLURM_ARRAY_TASK_ID.png
Read more about array jobs here: Job Arrays.
Priority¶
Jobs are assigned a priority which can affect how long it takes for them to be scheduled. This allows us to ensure that usage of Midway is fair and no one person or group is dominating the machine. Priority is calculated using Slurm’s multifactor plugin (http://slurm.schedmd.com/priority_multifactor.html), which takes into account these factors:
- AGE - this factor increases with the amount of time a job has been queued.
- FAIRSHARE - higher for users or groups who have not submitted jobs recently; decreases as machine use increases.
- JOBSIZE - higher for smaller jobs.
Fairshare priority is calculated per-user and per-group, so usage by other members of your group will decrease your job’s priority.
The priority of queued jobs can be determined with the sprio command.
View slurm estimated start times for pending jobs with this command:
squeue --start -p sandyb
Accounts and Allocations¶
RCC provides a handy tool for checking account information including the accounts you belong to, their balances, and cluster usage.
accounts options:
| Command | Description |
|---|---|
| accounts list | List the accounts the user belongs to |
| accounts members | List all members of accounts user belongs to |
| accounts allocations | List the active allocations for the user’s accounts |
| accounts storage | List storage allocations for the user’s accounts |
| accounts balance | Report the balance for the user’s accounts |
| accounts usage | Report the user’s usage in the current period |
| accounts partitions | List the partitions where usage is enforced (charged) |
| accounts periods | List current and past allocation periods |
The usage option also has a number of useful options for changing how usage
information is reported.
| Option | Description |
|---|---|
--accounts |
usage broken down by account |
--partitions |
usage broken down by partition |
--byperiod |
usage broken down by allocation period |
--bypartition |
usage broken down by by partition |
--byjob |
list usage by slurm job |
--all |
include partitions that aren’t charged |
Accessing Data¶
Samba¶
SAMBA allows uses to connect to (or “mount”) their home and project directories on their local computer so that the file system on Midway appears as if it were directly connected to the local machine. This method of accessing your RCC home and project space is only available from within the UChicago campus network. From off-campus you will need to first connect through the UChicago VPN.
Your SAMBA account credentials are your CNetID and password:
Username: ADLOCAL\CNetID
Password: CNet password
Hostname: midwaysmb.rcc.uchicago.edu
Note
Make sure to prefix your username with ADLOCAL\
See Data Transfer for platform specific details.
Globus Online¶
Globus Online is a robust tool for transferring large data files to/from Midway. The RCC has a customized Globus Online login site at https://globus.rcc.uchicago.edu and uses Single Sign On capabilities of CILogon. If you have already signed up, here is the connection information:
URL: https://globus.rcc.uchicago.edu
End Point: ucrcc#midway
You can also download a local client program for your laptop or desktop at https://www.globus.org/globus-connect-personal.
HTTP (web access)¶
RCC provides web access to data on their storage system via public_html
directories in both users’ home and project directories.
| Local path | Corresponding URL |
|---|---|
/home/drudd/public_html/research.dat |
http://users.rcc.uchicago.edu/~drudd/research.dat |
/project/rcc/public_html/results.txt |
http://projects.rcc.uchicago.edu/~rcc/results.txt |
Be sure your home or project directory and public_html have the execute bit set, and that public_html has read permissions if you would like to allow indexing (directory listings and automatic selection of index.html).
chmod o+x $HOME
mkdir -p $HOME/public_html
chmod o+rx $HOME/public_html
Files in public_html must also be readable by the web user (other), but should not be executable.
Note
Use of these directories must conform with the RCC usage policy (https://rcc.uchicago.edu/about-rcc/rcc-user-policy). Please notify RCC if you expect a large number of people to access data hosted here.
Specialized Environments¶
Python¶
Python Modules¶
RCC’s Python packages include a large number of sub-packages that are compiled and tuned for Midway. A list
of all packages and their versions can be found by loading the module and running the command pip list.
Since these packages are always under development, and are released on a variety of schedules, RCC provides a new Python module quarterly, including the most up-to-date versions of each package at that time. If you have specific version requirements that are not supplied by any of the RCC modules, please contact us for help.
The following table is a list of some of the largest and most popular libraries for the latest python module,
python/2.7-2015q2:
| Package | Version | Description |
|---|---|---|
| Numpy | 1.9.2 | Base N-dimensional array package |
| SciPy | 0.15.1 | Numerical routines for mathematics, science, and engineering. |
| Matplotlib | 1.4.3 | Plotting and visualization library |
| IPython | 3.1.0 | Interactive shell and browser-based notebook |
| Pandas | 0.16.0 | Data structures and analysis |
| scikit-learn | 0.16.0 | Machine learning and data mining |
| Sympy | 0.7.6 | Symbolic mathematics and computer algebra system. |
Installing python packages¶
Users may install their own python packages using pip, or by manually setting an installation
path to a setup.py script (if supported). If you install a python package into your home directory,
be sure to update or remove it if you change the system-wide python module as it may not be compatible.
pip install --user
These packages will be installed into $HOME/.local. You may need to add these directories to your
path if they include executables.
export PATH=$HOME/.local:$PATH
You can change which directory is used for local packages by defining the environment variable PYTHONUSERBASE.
Virtualenv¶
Python includes a mechanism for isolating packages into directories. Particularly useful for explicitly tracking
dependencies and required versions. Use the --system-site-packages option to include all RCC packages, which
have been linked to mkl and optimized for Midway.
$ module load python/2.7-2015q2
$ virtualenv --system-site-packages temp
New python executable in temp/bin/python
Installing setuptools, pip...done.
$ source temp/bin/activate
(temp)$ $ which pip
~/temp/bin/pip
Note
If you use the --system-site-packages flag you must load the same python RCC module and ensure compatibility
with locally installed packages in .local.
R¶
RStudio¶
RStudio is an IDE for programming in the R programming language. The “rstudio” modules on Midway will load the system default R module (R/2.15) if no R module is loaded. In general, load your favorite R module, then load RStudio, and then run it. For example, log into Thinlinc, then:
module load R/3.1 rstudio/0.99
rstudio
R packages¶
Similarly to the Python environment, RCC’s R modules include sub-packages developed by the R community. List all
available packages with the command installed.packages(). We will install or update packages upon request, provided
that they are published in the CRAN repository (http://cran.r-project.org/).
Installing R packages¶
Users may install their own R packages to their home folder by setting the R_LIBS_USER environment variable. For example:
export R_LIBS_USER=~/R_libs # path to folder for local R packages
mkdir -p $R_LIBS_USER # create this folder if it does not exist
module load R/3.1
R
> install.packages('mypackage')
Packages installed locally are only usable with the R version that they were installed with – so, in the example above, ‘mypackage’ can only be used with R/3.1. Be sure to set the R_LIBS_USER environment variable in order to make R aware of this package location, or put “export R_LIBS_USER=~/R_libs” into your ~/.bashrc file so it is executed every time you log in.
Matlab¶
Matlab Parallel Computing Toolbox, and in particular the matlab/2014b module, allows RCC users to easily
parallelize their Matlab calculations over a single Midway node.
Specific care must be taken when running multiple PCT jobs on Midway. The Matlab PCT requires a temporary “Job Storage Location” where it
stores information about the Matlab pool that is in use. This is simply a directory on the filesystem that Matlab writes various files to
in order to coordinate the parallelization of the matlabpool. By default, this information is stored in /home/YourUsername/.matlab/, which
can only be used by a single running job at a time.
The solution is to have each of your jobs that will use the PCT set a unique location for storing job information. To do this, a temporary directory must be created before launching matlab in your submission script and then the matlabpool must be created to explicitly use this unique temporary directory. Example sbatch and matlab scripts are shown below:
#!/bin/bash
#SBATCH --job-name=whatever
#SBATCH --output=matlab_parfor.out
#SBATCH --error=matlab_parfor.err
#SBATCH --partition=sandyb
#SBATCH --time=00:10:00
#SBATCH --nodes=1
#SBATCH --ntasks=16
module load matlab/2014b
# Create a local work directory
mkdir -p /tmp/YourUsername/$SLURM_JOB_ID
# Kick off matlab
matlab -nodisplay < multi_parfor.m
# Cleanup local work directory
rm -rf /tmp/YourUsername/$SLURM_JOB_ID
The most basic level of parallelization in Matlab is achieved through use of a parfor loop in place of a for loop. The iterations of a parfor loop are distributed to the workers in the active matlabpool and computed concurrently. For this reason, care must be taken to ensure that each iteration of the parfor loop is independent of every other.
% create a local cluster object
pc = parcluster('local')
% explicitly set the JobStorageLocation to the temp directory that was created in your sbatch script
pc.JobStorageLocation = strcat('/tmp/YourUsername/', getenv('SLURM_JOB_ID'))
% start the matlabpool with maximum available workers
% control how many workers by setting ntasks in your sbatch script
matlabpool(pc, getenv('SLURM_CPUS_ON_NODE'))
% run a parallel for loop
parfor i = 1:100
ones(10,10)
end
Spark¶
Spark is a framework for large-scale data processing that is supplanting Hadoop for some applications in industry. Spark’s Python API (“pyspark”) is a layer over the Python shell that allows you to do distributed computing using the Python constructs you already know and love. For example:
- Reserve an exclusive node:
sinteractive --exclusive
- Load the spark and python modules, and then start a pyspark shell.
module load spark python/2.7-2015q2
pyspark
3) Code away! This example takes a text file that represents a matrix and returns the number of rows in which the row sum is greater than 100.
myfile = sc.textFile("numbers.txt")
myfile.map(lambda line: sum([int(x) for x in line.strip().split(' ')])).filter(lambda x: x>100).count()
Pyspark can also be used in Slurm scripts, or with multinode jobs. See more at Spark.
Debugging and Optimization¶
perf¶
Perf is a command line profiling tool useful for obtaining a high-level view of a program’s performance characteristics. It uses hardware counters to monitor cache misses, instructions performed, branches and branch- misses, and other factors that influence performance. It is available on Midway without a module (as part of the system installation).
$ perf stat ./a.out
Time using 32 threads = 0.811506
Performance counter stats for './a.out':
36401.777859 task-clock # 3.210 CPUs utilized
3,558 context-switches # 0.098 K/sec
151 cpu-migrations # 0.004 K/sec
3,906,521 page-faults # 0.107 M/sec
109,932,188,744 cycles # 3.020 GHz [83.35%]
86,296,015,582 stalled-cycles-frontend # 78.50% frontend cycles idle [83.37%]
65,727,376,387 stalled-cycles-backend # 59.79% backend cycles idle [66.66%]
59,984,433,422 instructions # 0.55 insns per cycle
# 1.44 stalled cycles per insn [83.31%]
6,058,066,960 branches # 166.422 M/sec [83.34%]
4,723,714 branch-misses # 0.08% of all branches [83.29%]
11.339093418 seconds time elapsed
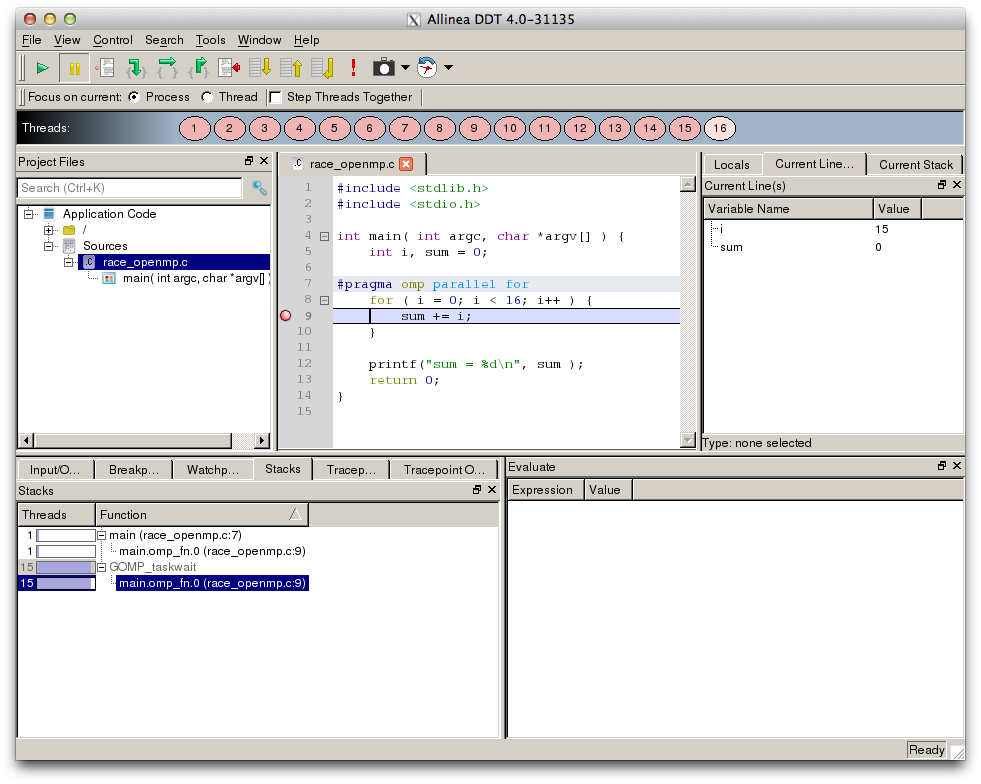
Allinea ddt¶
Allinea’s DDT (Distributed Debugging Tool) is a powerful, commercial gui-based debugger used- in many HPC environments for debugging large MPI and OpenMP parallel programs. For more information see Allinea DDT.
Compiler Optimization¶
RCC Midway contains nodes spanning 3 generations of Intel processors and one generation of AMD processor. Unless you specify otherwise, compilers will optimize for the processor generation of the compiling node, in this case the login nodes. If you expect to run your code on nodes with processors other than SandyBridge, you should specify the compilation target architectures for best performance.
| Partition | Processor Generation | Optimization flags |
|---|---|---|
| westmere | Intel Xeon X5675 | -xSSE4.2 (intel), -march=westmere (gcc), -tp nehalem (pgi) |
| sandyb | Intel Xeon E5-2670 | -xAVX (intel), -march=sandybridge (gcc), -tp sandybridge (pgi) |
| ivyb | Intel Xeon E5-2680 | -xAVX (intel), -march=ivybridge (gcc), -tp ivybridge (pgi) |
| amd | AMD Opteron 6386 SE | -march=bdver2 (gcc), -tp piledriver (pgi) |
Additionally, Intel and PGI support compiling for multiple targets, which can be selected at runtime. To compile your code to be optimized for all Midway architectures, use the following flags:
| Compiler Module | Flags |
|---|---|
| pgi/2013 | -tp nehalem,ivybridge,sandybridge,piledriver |
| intel/2015 | -axAVX,SSE4.2 |
| gcc/4.9 | -mtune=sandybridge -march=westmere |
Note
Using the intel compiler with the amd partition is not recommended. Instead, use GNU or PGI compiler suites.